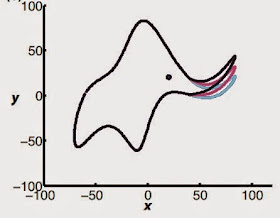
 |
| "With four parameters you can fit an elephant to a curve, with five you can make him wiggle his trunk" - John von Neumann |
The nice thing about Derek's talk was that it was really delivered from the other side of the fence, that of an accomplished and practicing medicinal chemist. Thus he wisely did not dwell too much on all the details that can go wrong in modeling: since the audience mainly consisted of modelers presumably they knew these already (No, we still can't model water well. Stop annoying us!). Instead he offered a more impressionistic and general perspective informed by experience.
Why von Neumann's elephants? Derek was referring to a great piece by Freeman Dyson (who I have had the privilege of having lunch with a few times) published in Nature a few years back in which Dyson reminisced about a meeting with Enrico Fermi in Chicago. Dyson had taken the Greyhound bus from Cornell to tell Fermi about his latest results concerning meson-proton scattering. Fermi took one look at Dyson's graph and basically demolished the thinking that had permeated Dyson and his students' research for several years. The problem, as Fermi pointed out, was that Dyson's premise was based on fitting a curve to some data using four parameters. But, quipped Fermi, you can get a better fit to the data if you add more parameters. Fermi quoted his friend, the great mathematician John von Neumann - "With four parameters you can fit an elephant to a curve; with five you can make him wiggle his trunk". The conversation lasted about ten minutes. Dyson took the next bus back to Ithaca.
And making elephants dance is indeed what modeling in drug discovery runs the risk of doing, especially when you keep on adding parameters to improve the fit (Ah, the pleasures of the Internet - turns out you can literally fit an elephant to a curve). This applies to all models, whether they deal with docking, molecular dynamics or cheminformatics. This problem of overfitting is well-recognized, but researchers don't always run the right tests to get rid of it. As Derek pointed out however, the problem is certainly not unique to drug discovery. He started out by describing the presence of "rare" events in finance related to currency fluctuations - these rare events happen often enough and their magnitude is devastating enough to cause major damage. Yet the models never captured them, and this failure was responsible at least in part for the financial collapse of 2008 (this is well-documented in Nassim Taleb's "The Black Swan").
Here are the charges brought against modelers by medicinal chemists: They cannot always predict but often can only retrodict, they cannot account for 'Black Swans' like dissimilar binding modes resulting from small changes in structure, they often equate computing power with accuracy or confidence in their models, they promise too much and underdeliver. These charges are true to varying degrees under different circumstances, but it's also true that most modelers worth their salt are quite aware of the caveats. As Derek pointed out with some examples however, modeling has over-promised for a long time: a particularly grating example was this paper from Angewandte Chemie from 1986 in which predicting the interaction between a protein and small molecule is regarded as a "spatial problem" (energy be damned). Then there's of course the (in)famous "Designing Drugs Without Chemicals" piece from Fortune magazine which I had highlighted on Twitter a while ago. These are all good examples of how modeling has promised riches and delivered rags.
However I don't think modeling is really any different from the dreams spun from incomplete successes by structure-based design, high-throughput screening, combinatorial chemistry or efficiency metrics. Most of these ideas consist of undue excitement engendered by a few good successes and inflated dreams of what's sometimes called "physics envy" - the idea that your discipline holds the potential to become as accurate as atomic physics if only everyone adopted your viewpoint. My feeling is that because chemistry unlike physics is primarily an experimental discipline, chemists are generally more inherently skeptical of theory, even when it's doing no worse than experiment. In some sense the charge against modeling is a bit unfair since it's also worth remembering that unlike synthetic organic chemistry or even biochemistry which have had a hundred and fifty years to mature and hone themselves into (moderately) predictive sciences, the kind of modeling that we are talking about is only about three decades old. Scientific progress does not happen overnight.
The discussion of hype and failure brings us to another key part of Derek's talk, that of recognizing false patterns in the data and getting carried away by their apparent elegance, sophisticated computing power or just plain random occasional success. This part is really about human psychology rather than science so it's worth noting the names of psychologists like Daniel Kahneman and Michael Shermer who have explored human fallibility in this regard. Derek gave the example of the "Magic Tortilla" which was a half-baked tortilla in which a lady from New Mexico saw Jesus's image - the tortilla is now in a shrine in New Mexico. In this context I would strongly recommend Michael Shermer's book "The Believing Brain" in which he points out human beings' tendency to see false signals in a sea of noise - generally speaking we are much more prone to seeing false positives rather than false negatives, and this kind of Type I error makes sense when we think about how false positives would have saved us from menacing beasts on the prairie while false negatives would simply have weeded us out of the gene pool. As Derek quipped, the tendency to see signal in the data has basically been responsible for much scientific progress, but it can also contribute mightily to what Irving Langmuir called "pathological science". Modeling is no different - it's very easy to extrapolate from a limited number of results and assume that your success applies to data that is far more extensive and dissimilar to what your methods have been applied to.
There are also some important questions here about what exactly what it is that computational chemists should suggest to medicinal chemists, and much of that discussion arose in the Q&A session. One of the points that was raised was that modelers should stick their necks out and should make non-intuitive suggestions - for instance simply asking a medicinal chemist to put a fluorine or methyl group on an aromatic ring is not as useful because the medicinal chemist might have thought of that as a random perturbation anyway. However this suggestion is not as easy as it sounds since the largest changes are also often the most uncertain, so at the very least it's the modeler's responsibility to communicate this level of risk to his or her colleagues. As an aside, this reminds me of Karl Popper since one of the criteria Popper used for distinguishing a truly falsifiable and robust theory from the rest was its ability to stick its neck out and make bold predictions. As a modeler I also think it's really important to realize when to be coarse-grained and when to be fine-grained. Making very precise and limited suggestions when you don't have enough data and accuracy to be fine-grained is asking for trouble, so that's a decision you should be constantly making throughout the flow of a project.
One questioner also asked if as a medicinal chemist Derek would make dead compounds to test a model - this is a question that is very important in my opinion since sometimes negative suggestions can provide the best interrogation of a model. I think the answer to this question is best given on a case-by-case basis: if the synthetic reaction is easy and can rapidly generate analogs then the medicinal chemist should be open to the suggestion. If the synthesis is really hard then there should be a damn good reason for doing it, often a reason that would lead to a crucial decision in the project. The same goes for positive suggestions too: a positive suggestion that may potentially lead to significant improvement should be seriously considered by the medicinal chemist, even if it may be more uncertain than one which is rock-solid but only leads to a marginal improvement.
Derek ended with some sensible exhortations for computational chemists: Don't create von Neumann's elephants by overfitting the data, don't talk about what hardware you used (effectively giving the impression that just because you used the latest GPU-based exacycle your results must be right: and really, the medicinal chemist doesn't care), don't see patterns in the data (magic tortillas) based on limited and spotty results.
Coupled with these caveats were more constructive suggestions: always communicate the domain of applicability/usability of your technique and the uncertainty inherent in the protocol, realize that most medicinal chemists are clueless about the gory details of your algorithm and therefore will take what you say at face value (and this also goes to show how it's your responsibility as modelers to properly educate medicinal chemists), admit failures when they exist and try to have the failures as visible as the successes (harder when management demands six sigma-sculpted results but still doable). And yes, it never, ever hurts for modelers to know something about organic chemistry, synthetic routes and the stability of molecules. I would also add the great importance of using statistics to analyze and judge your data.
All very good suggestions indeed. I end with only one: realize that you are all in this together and that you are all - whether modelers, medicinal chemists or biologists - imperfect minds trying to understand highly complex biological systems. In that understanding will be found the seeds of (occasional) success.
Added: It looks like there's going to be a YouTube video of the event up soon. Will link to it once I hear more.
Excellent post, Ash, which does not betray the timelines that you were working to. A most diligent wavefunction! Something that drug discovery scientists need to think about is whether pharmaceutical computational chemistry focuses too much on prediction and not enough on design.
ReplyDeleteThanks Pete. Yes, good point: molecular design is definitely an important part of what we do, and it's an aspect in which both computational and medicinal chemists can work together. One thing I forgot to note is that we never really strayed into things like similarity searching and cheminformatics due to time constraints and stuck mainly to what we call "physics-based" modeling. As you know those things are a big part of computational chemistry, although they too are plagued by similar problems.
DeleteThis is relevant and perhaps of interest to readers: http://www.lassp.cornell.edu/sethna/Sloppy/
ReplyDelete